Low Coefficients of Variation from Run-to-Run when Enriching Proteins from Human Plasma Samples
- May 27, 2025
- 1 min read
In proteomics, achieving high reproducibility and deep proteome coverage is essential for confident biomarker discovery, especially when working with complex biological samples like human plasma. One of the most critical metrics for assessing reproducibility is the coefficient of variation (CV), which quantifies the consistency of protein quantification across replicate runs. Lower CVs translate to greater confidence in detected differences and reduce the number of replicates needed to achieve statistical significance, saving both time and resources.(1)
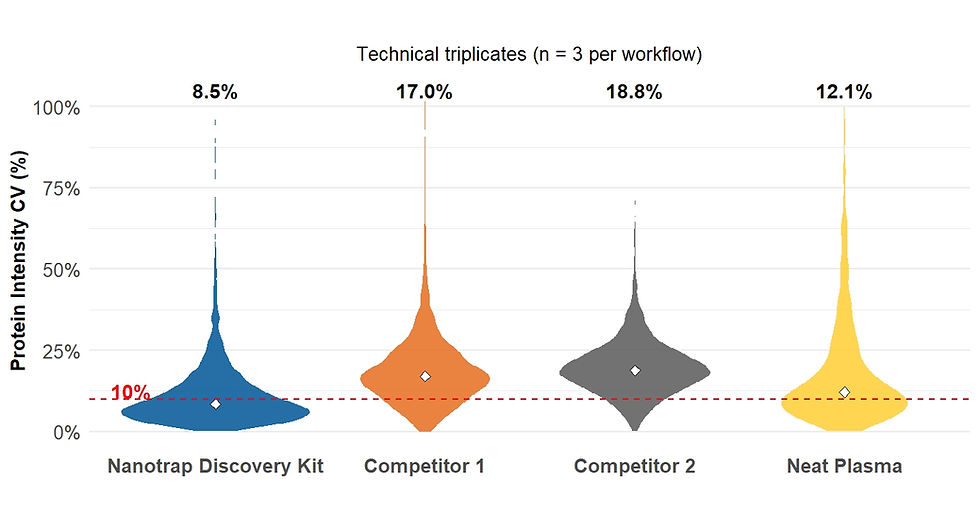
In this study, we demonstrate that the Nanotrap® Protein Enrichment Affinity Kit (PEAK)—applied through both manual and semi-automated protocols—enables substantial improvements in both proteome coverage and technical reproducibility.(2,3) Compared to neat plasma processing, the Nanotrap PEAK manual and KingFisher™ Apex System protocols reduced median protein CVs from 13% to 8.4% and 2.3%, respectively. This reduction represents up to a 32-fold decrease in the number of replicates required to achieve equivalent statistical power. These results establish Nanotrap PEAK workflows as highly efficient and reproducible solutions for plasma proteomics, supporting both deep discovery and robust quantitation.

APPLICATION NOTE SKU # 34XXX
Lit. # PL-AN31416


Comments